Table of Links
Abstract and I. Introduction
-
Materials and Methods
2.1. Multiple Instance Learning
2.2. Model Architectures
-
Results
3.1. Training Methods
3.2. Datasets
3.3. WSI Preprocessing Pipeline
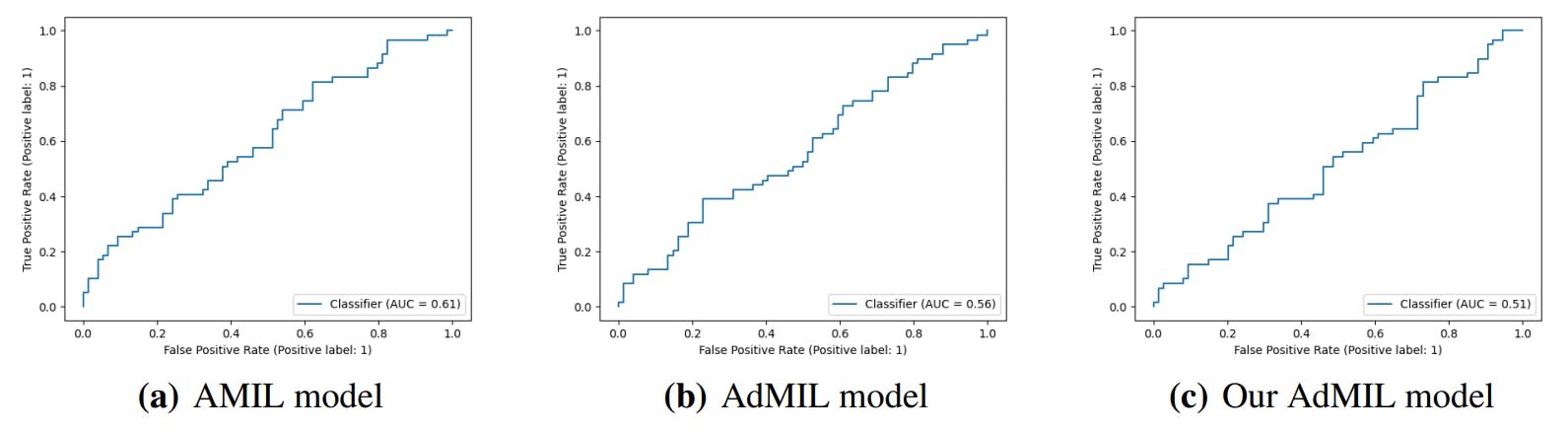
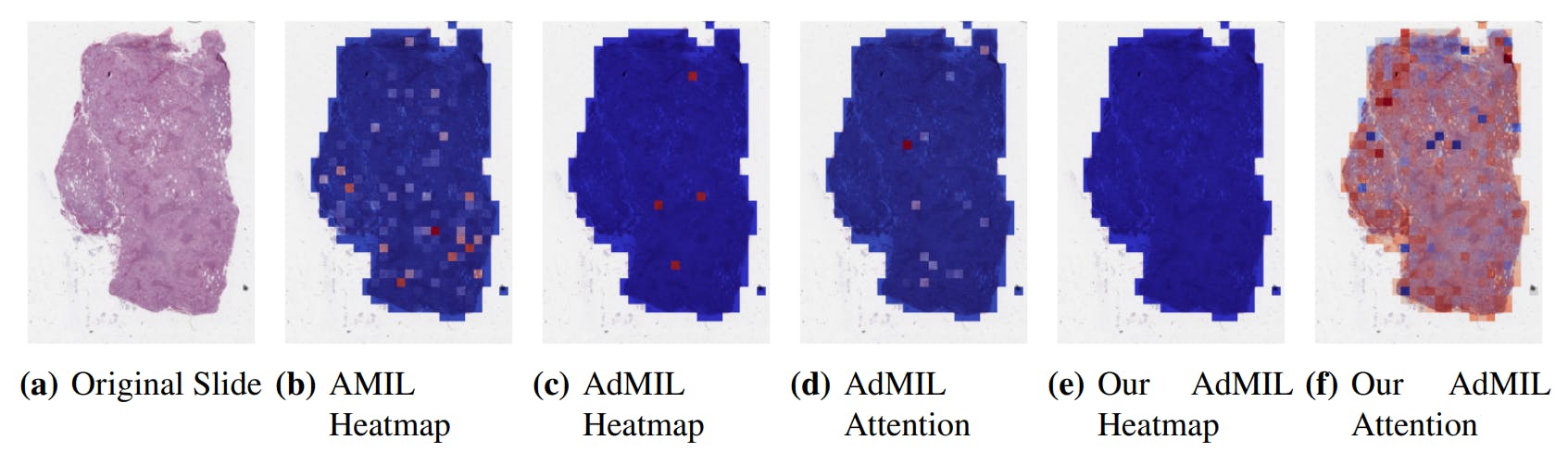
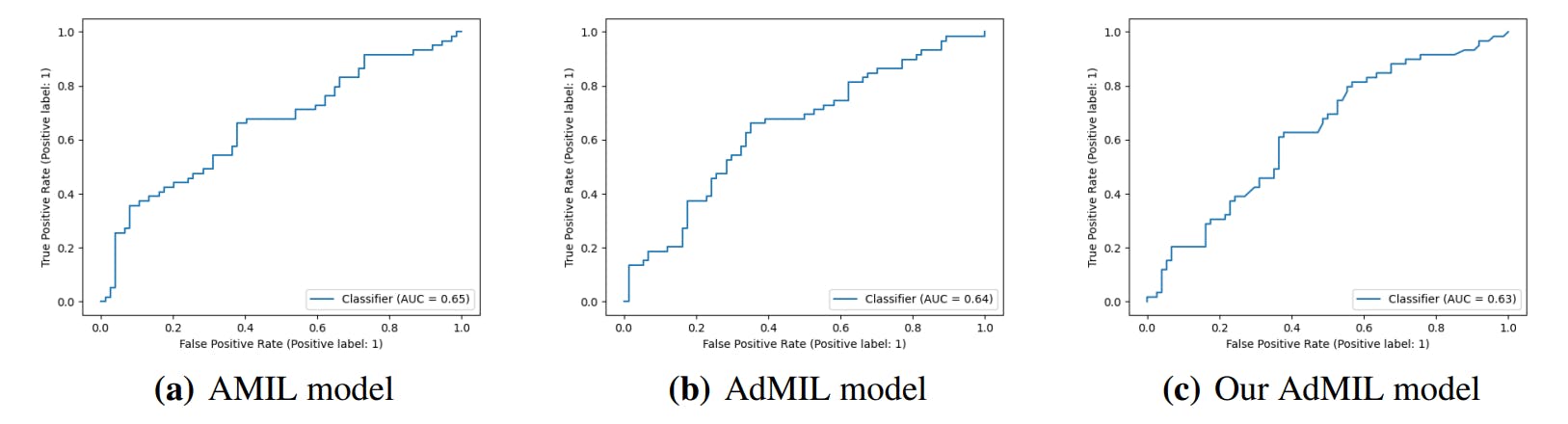
3.4. Classification and RoI Detection Results
-
Discussion
4.1. Tumor Detection Task
4.2. Gene Mutation Detection Task
-
Conclusions
-
Acknowledgements
-
Author Declaration and References
3.3. WSI Preprocessing Pipeline
WSIs are often too large to be directly processed by Deep Learning models. Slides can occupy over 6 GB of memory, so having a dataset of raw slides on disk is not feasible in our case. To store our dataset locally, we decided to focus on one magnification at a time. All our models were trained in separate magnification levels, which guaranteed that this would not be an issue. For some magnifications, the total of patches might exceed the storage space available. To overcome this issue, we developed a pipeline that fetches WSI tiles, processes, filters, efficiently encodes them, and saves the resulting embeddings and relevant metadata on HDF5 files. This pipeline is explained in the following sections.
3.3.1. WSI Metadata Extraction
We start by extracting information about each slide: id, labels type of slide, and microns per pixel (mpp) at which the slide was originally scanned. In the end, all the metadata extracted is saved to a CSV (comma-separated values) file for further processing.
3.3.2. Patch Fetching and Pre-processing
Whole slide images can have a lot of patches consisting exclusively of background or artifacts that make them unusable for training our models. Fetching all these unnecessary patches and checking them one by one becomes inefficient and time-consuming. To avoid this we take advantage of WSI’s multi-scale property to avoid fetching patches that will definitely contain only background.
The tile at the thumbnail level is initially fetched. Otsu’s thresholding [16] is applied to it, as well as a close morphological operation to filter noise. We then extract the resulting black pixel coordinates, obtaining a set of pixels 𝑃 , the pixels that correspond to tissue.
In the acquisition of a WSI, there are often unintentional artifacts due to manual tissue preparation, staining, and scanning hardware, as well as pathologists’ annotations. To mitigate the number of tiles containing artifacts as well as remove some background pixels that might have passed through the previous filter, the color of each pixel 𝑝 ∈ 𝑃 was compared to the average color of 𝑃 , by calculating their Euclidean distance and comparing it with a pre-defined threshold. The coordinates of the pixels that fulfilled this condition were then stored and used to calculate the corresponding coordinates of the patches at the desired magnification, using the hierarchical properties of WSI and the metadata extracted from the step in 3.3.1.
Because these filters were applied at the thumbnail level, some of the tiles were still not suitable for the final dataset. After fetching each tile, we checked if its size in image coordinates was 512 × 512 px. If it was smaller, it was padded accordingly with the average background color. The percentage of tissue present in the tile was then calculated and compared with a threshold. If it does not contain enough tissue, the image is discarded.
For Gene Mutation Detection, due to the size of the FFPE slides and the multiple magnification levels used, we applied random sampling to the tiles:
• At 5x magnification, we sampled 60% of the filtered tiles, when their number was greater than a certain limit; otherwise, we used all the tiles.
• In the case of 10x and 20x magnification, we applied clustering on the tiles from the previous magnification and sampled a chosen number n of tiles per cluster. We then proceeded to use the hierarchical properties of WSI to extract the corresponding tiles at the desired magnifications. For instance, we performed K-means clustering on the tiles at 5x magnification, sampled a maximum of 20 tiles from each cluster, and proceeded to extract, for each tile, the corresponding tiles at 10x magnification (Figure 2).
3.3.3. Feature Extraction
As mentioned previously, embeddings were created from the tiles. For this, we chose KimiaNet[18], a Densenet 121 pretrained for WSI tumor subtype classification, that produces embedding vectors with a length of 1024. This model was trained exclusively on FFPE slides.
We also performed two data augmentations per tile, composed of random HED stain perturbation, Gaussian noise addition, rotations, and horizontal and vertical flips. Embeddings were generated for these augmented tiles as well. The embeddings are then saved to an HDF5 file, along with the corresponding relevant metadata, such as the coordinates of the patches and labels. This file can then be quickly read to memory during the training process of the models.
Due to the size of the dataset, immediately converting each 512 × 512 pixels patch to an embedding of length 1024 saves storage space (with a decrease in size close to a thousand-fold) and time for training the model and allows us to have the whole dataset locally, instead of having to fetch the data each time we needed to train or fine-tune the models. Furthermore, by having the slides represented in feature space immediately, as opposed to pixel space, we were able to fit all patches in a slide into GPU memory concurrently, which is especially useful for multiple instance learning approaches.
Authors:
(1) Martim Afonso, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, Lisbon, 1049-001, Portugal;
(2) Praphulla M.S. Bhawsar, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(3) Monjoy Saha, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(4) Jonas S. Almeida, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(5) Arlindo L. Oliveira, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, Lisbon, 1049-001, Portugal and INESC-ID, R. Alves Redol 9, Lisbon, 1000-029, Portugal.
This paper is