Table of Links
Abstract and I. Introduction
-
Materials and Methods
2.1. Multiple Instance Learning
2.2. Model Architectures
-
Results
3.1. Training Methods
3.2. Datasets
3.3. WSI Preprocessing Pipeline
3.4. Classification and RoI Detection Results
-
Discussion
4.1. Tumor Detection Task
4.2. Gene Mutation Detection Task
-
Conclusions
-
Acknowledgements
-
Author Declaration and References
5. Conclusions
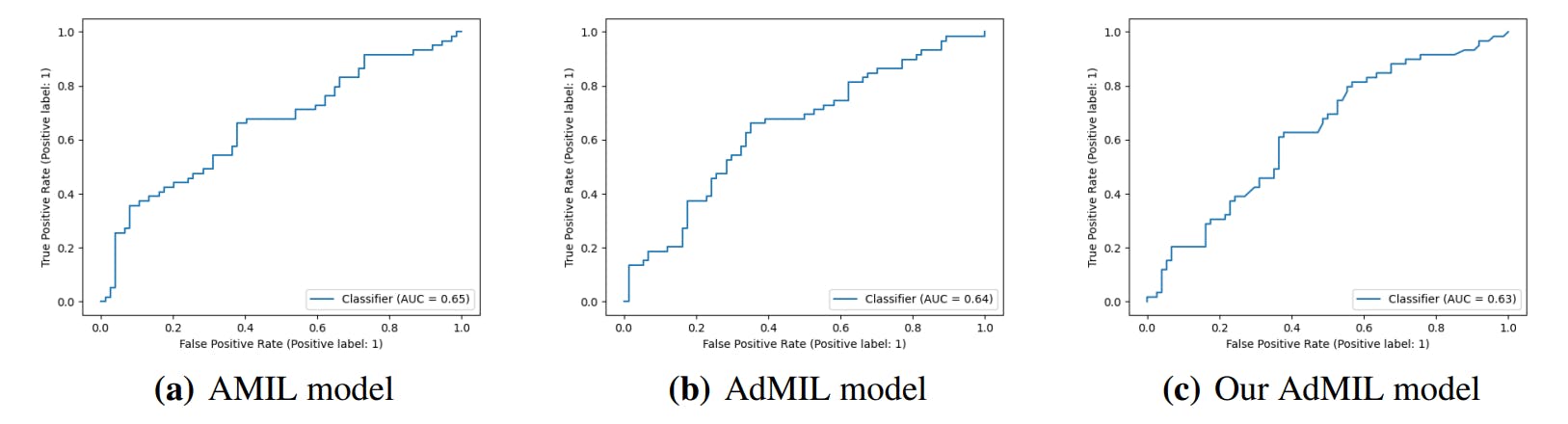
In this work, we study the performance of multi-instance learning (MIL) frameworks with attention mechanisms for WSI classification and virtual staining of breast tumors. We explored two distinct approaches that identify different morphology proxies for patch (tile) classifiers. These two frameworks were used in a weakly-supervised application to tumor detection and TP53 mutation detection in Breast Carcinoma Lung and Squamous Cell Carcinoma. We found that it was far easier to identify Regions of Interest (RoIs) recognizing tumor vs non-tumor even at low resolution (AUC > 0.95), than it was to classify TP53 mutated vs non-mutated (AUC < 0.71). The observation that higher resolutions (20x) worked better to identify RoI for mutation was by itself not a surprise, but the opportunities to hypothesize novel morphological interpretations emerged as the main result from the work reported here. Similarly, when we explored new modifications of the established multi-instance learning method by altering its original attention layer, the most interesting result was not improved accuracy but the attention the model placed on different morphological features ( i.e. “slide painting”). Specifically, the results described in Figures 3, 4, and 5 illustrate the opportunities for interactive exploration of recurring morphologies for their role in cancer etiology.
6. Acknowledgements
This work was funded in part by the National Cancer Institute (NCI) Intramural Research Program CAS#10901 (EpiSphere), the Recovery and Resilience Fund towards the Center for Responsible AI (Ref. C628696807-00454142), and the financing of the Foundation for Science and Technology (FCT) for INESCID (Ref. UIDB/50021/2020).
The open source code used in this project is publicly available at https://github.com/timafonso/WSI_MIL_ROI
7. Author Declaration
There is no conflict of interest to report.
References
[1] Bahdanau, D., Cho, K.H., Bengio, Y., 2015. Neural machine translation by jointly learning to align and translate. 3rd International Conference on Learning Representations, ICLR 2015 – Conference Track Proceedings , 1–15.
[2] Dietterich, T.G., Lathrop, R.H., Lozano-Pérez, T., 1997. Solving the multiple instance problem with axis-parallel rectangles. Artificial Intelligence 89, 31–71.
[3] Dimitriou, N., Arandjelović, O., Caie, P.D., 2019. Deep Learning for Whole Slide Image Analysis: An Overview. Frontiers in Medicine 6, 1–7.
[4] Guo, B., Li, X., Yang, M., Zhang, H., Xu, X.S., 2023. A robust and lightweight deep attention multiple instance learning algorithm for predicting genetic alterations. Computerized Medical Imaging and Graphics 105, 102189.
[5] Hanna, M.G., Parwani, A., Sirintrapun, S.J., 2020. Whole slide imaging: technology and applications. Advances in Anatomic Pathology 27, 251–259.
[6] Hou, W., Yu, L., Lin, C., Huang, H., Yu, R., Qin, J., Wang, L., 2022. H2-MIL: Exploring Hierarchical Representation with Heterogeneous ˆ Multiple Instance Learning for Whole Slide Image Analysis. Proceedings of the AAAI Conference on Artificial Intelligence 36, 933–941.
[7] Ilse, M., Tomczak, J., Welling, M., 2018. Attention-based deep multiple instance learning, in: International conference on machine learning, pp. 2127–2136.
[8] Javed, S.A., Juyal, D., Padigela, H., Taylor-Weiner, A., Yu, L., Prakash, A., 2022. Additive mil: Intrinsically interpretable multiple instance learning for pathology. Advances in Neural Information Processing Systems 35, 20689–20702.
[9] Khened, M., Kori, A., Rajkumar, H., Krishnamurthi, G., Srinivasan, B., 2021. A generalized deep learning framework for whole-slide image segmentation and analysis. Scientific reports 11, 1–14.
[10] Konstantinov, A.V., Utkin, L.V., 2022. Multi-attention multiple instance learning. Neural Computing and Applications , 1–23.
[11] Li, B., Li, Y., Eliceiri, K.W., 2021. Dual-stream Multiple Instance Learning Network for Whole Slide Image Classification with Self-supervised Contrastive Learning. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition , 14313–14323.
[12] Li, Z., Yuan, L., Xu, H., Cheng, R., Wen, X., 2020. Deep Multi-Instance Learning with Induced Self-Attention for Medical Image Classification. Proceedings – 2020 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2020 , 446–450.
[13] Lu, M., Pan, Y., Nie, D., Liu, F., Shi, F., Xia, Y., Shen, D., 2021a. Smile: Sparse-attention based multiple instance contrastive learning for glioma sub-type classification using pathological images, in: MICCAI Workshop on Computational Pathology, pp. 159–169.
[14] Lu, M.Y., Williamson, D.F., Chen, T.Y., Chen, R.J., Barbieri, M., Mahmood, F., 2021b. Data-efficient and weakly supervised computational pathology on whole-slide images. Nature Biomedical Engineering 5, 555–570.
[15] Nayak, N., Chang, H., Borowsky, A., Spellman, P., Parvin, B., 2013. Classification of tumor histopathology via sparse feature learning, in: 2013 IEEE 10th international symposium on biomedical imaging, pp. 410–413.
[16] Otsu, N., 1979. A threshold selection method from gray-level histograms. IEEE transactions on systems, man, and cybernetics 9, 62–66.
[17] Reisenbüchler, D., Wagner, S.J., Boxberg, M., Peng, T., 2022. Local attention graph-based transformer for multi-target genetic alteration prediction, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 377–386.
[18] Riasatian, A., Babaie, M., Maleki, D., Kalra, S., Valipour, M., Hemati, S., Zaveri, M., Safarpoor, A., Shafiei, S., Afshari, M., Rasoolijaberi, M., Sikaroudi, M., Adnan, M., Shah, S., Choi, C., Damaskinos, S., Campbell, C.J., Diamandis, P., Pantanowitz, L., Kashani, H., Ghodsi, A., Tizhoosh, H., 2021. Fine-tuning and training of densenet for histopathology image representation using tcga diagnostic slides. Medical Image Analysis 70, 102032.
[19] Rymarczyk, D., Borowa, A., Tabor, J., Zielinski, B., 2021. Kernel self-attention for weakly-supervised image classification using deep multiple instance learning. Proceedings – 2021 IEEE Winter Conference on Applications of Computer Vision, WACV 2021 , 1720–1729.
[20] Shao, Z., Bian, H., Chen, Y., Wang, Y., Zhang, J., Ji, X., et al., 2021. Transmil: Transformer based correlated multiple instance learning for whole slide image classification. Advances in neural information processing systems 34, 2136–2147.
[21] Sheikh, T.S., Kim, J.Y., Shim, J., Cho, M., 2022. Unsupervised learning based on multiple descriptors for wsis diagnosis. Diagnostics 12, 1480.
[22] Tomczak, K., Czerwińska, P., Wiznerowicz, M., 2015. The cancer genome atlas (tcga): An immeasurable source of knowledge. Wspolczesna Onkologia 1A, A68–A77.
[23] Wang, X., Chen, H., Gan, C., Lin, H., Dou, Q., Tsougenis, E., Huang, Q., Cai, M., Heng, P.A., 2020. Weakly Supervised Deep Learning for Whole Slide Lung Cancer Image Analysis. IEEE Transactions on Cybernetics 50, 3950–3962.
[24] Wang, X., Zou, C., Zhang, Y., Li, X., Wang, C., Ke, F., Chen, J., Wang, W., Wang, D., Xu, X., et al., 2021. Prediction of brca gene mutation in breast cancer based on deep learning and histopathology images. Frontiers in Genetics 12, 661109.
[25] Yao, J., Zhu, X., Jonnagaddala, J., Hawkins, N., Huang, J., 2020. Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Medical Image Analysis 65.
[26] Zhao, L., Yuan, L., Hao, K., Wen, X., 2022. Generalized attention-based deep multi-instance learning. Multimedia Systems , 1–13.
[27] Zhu, X., Yao, J., Zhu, F., Huang, J., 2017. WSISA: Making survival prediction from whole slide histopathological images. Proceedings – 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017 2017-Janua, 6855–6863.
Authors:
(1) Martim Afonso, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, Lisbon, 1049-001, Portugal;
(2) Praphulla M.S. Bhawsar, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(3) Monjoy Saha, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(4) Jonas S. Almeida, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(5) Arlindo L. Oliveira, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, Lisbon, 1049-001, Portugal and INESC-ID, R. Alves Redol 9, Lisbon, 1000-029, Portugal.
This paper is