Table of Links
Abstract and I. Introduction
-
Materials and Methods
2.1. Multiple Instance Learning
2.2. Model Architectures
-
Results
3.1. Training Methods
3.2. Datasets
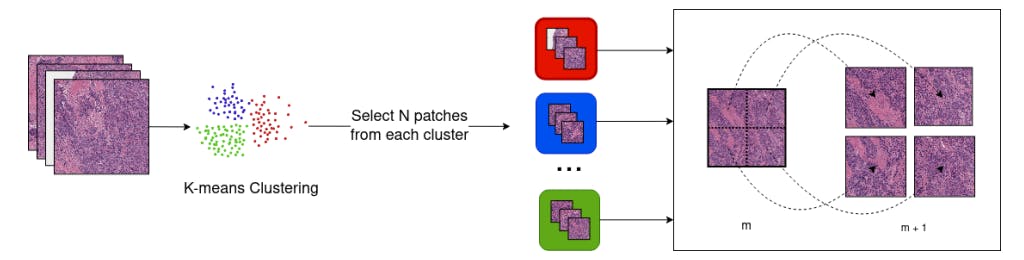
3.3. WSI Preprocessing Pipeline
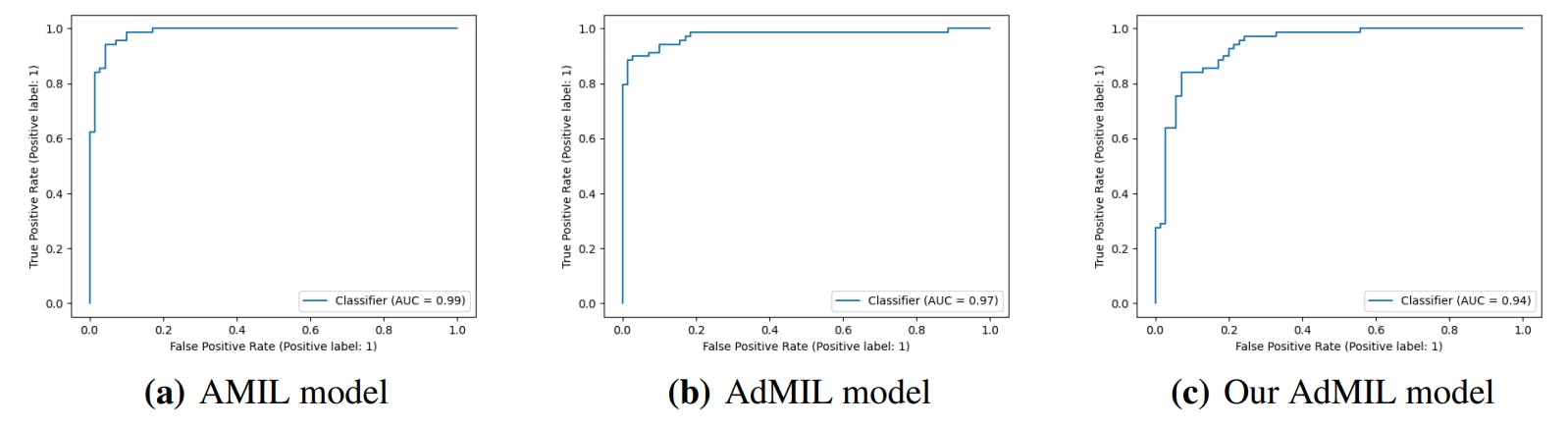
3.4. Classification and RoI Detection Results
-
Discussion
4.1. Tumor Detection Task
4.2. Gene Mutation Detection Task
-
Conclusions
-
Acknowledgements
-
Author Declaration and References
2.2. Model Architectures
Due to the need for interpretability of the results, when choosing and building the models, we focused on MIL models that use an attention pooling mechanism. Since we also prioritized efficiency, we chose models with a relatively simple architecture. Specifically, we focused on the original attention MIL (AMIL), proposed by Ilse et al. [7] and the additive MIL (AdMIL), proposed by Javed et al. [8].
Ilse et al. proposed a weighted average of the instances as a MIL pooling operator, where the weights were trained on a two-layered neural network. This operator corresponds to a version of the attention mechanism where all instances are independent.
In this approach, we have a 3-part model 𝑔, defined as
𝑎 is the attention module, defined as:
and 𝑝 is the predictor function or bag classifier:
Both 𝜓𝑎 and 𝜓𝑝 are multi-layer perceptrons.
The attention mechanism is similar to Badhanau’s attention [1], with the main difference that the instances are considered sequentially independent:
with both 𝑤 and 𝑉 as trainable parameters.
However, this approach still presents some problems, regarding what is being interpreted in the image, because the attention scores might not be expressive enough for the explainability of the results. Javed et al. [8] noted that the non-linear relationship between the attention values and the final prediction makes the visual interpretation inexact and incomplete.
The argument is that the attention scores obtained are not necessarily representative of tumors, but might still be needed for the downstream prediction, meaning they might be necessary but not sufficient for the final classification. The scores might indicate that the patch provides strong positive or negative evidence for the final prediction, but it does not make a distinction between the two. Moreover, the attention scores do not consider the contribution of multiple patches. Two patches might have moderate scores, but together they might provide strong evidence for the final prediction.
To address these problems, Ilse et al. proposed a new additive method for better visual interpretability and patch classification. To better specify the contributions from each patch, the final prediction function of the model described in equation 5 undergoes the following change:
where 𝜓𝑝 produces a score for each patch. With this change, 𝜓𝑝 (𝑎𝑖 (𝑥)) becomes the contribution for patch 𝑖 in a bag, turning it into a more accurate proxy of an instance classifier. The patch scores are then added to produce a final bag-level score, which in turn is converted to a final probability distribution using a 𝑠𝑜𝑓 𝑡𝑚𝑎𝑥, to obtain the final classification.
For interpretability and slide inpainting, the patch scores obtained can be passed through a 𝑠𝑖𝑔𝑚𝑜𝑖𝑑 function to produce a bounded patch contribution score between 0 and 1, where 0 – 0.5 values represent inhibitory scores and 0.5 – 1 values represent excitatory scores.
AMIL was originally trained with each WSI patch as bag (where each instance is a portion of that patch) for tumor detection, while AdMIL was trained for cancer sub-type classification and metastasis detection. The application of these models for tasks such as tumor detection and gene mutation detection can give provide insights into the models’ ability to generalize for different medical tasks. Furthermore, the differences in their employment of the MIL framework make it worthwhile to study and understand how these models focus on the different regions of slides.
The models’ original architectures used in this work are represented in Figure 1. The original AMIL model is composed of a feature extractor module, followed by the attention module which produces attention scores for each instance, and the final bag classifier module which takes the sum of all instances multiplied by their respective scores and outputs the final bag classification. Similarly to the AMIL architecture, the AdMIL model is composed of a feature extractor module and an attention module. However, as mentioned before, instead of aggregating the patch embeddings through a weighted sum and then passing the result through a final bag classifier, AdMIL passes each patch embedding multiplied by its attention score through a patch classifier, the Patch Score Layer. This classifier produces a patch score for each class. These scores are not bounded by any values and can be either negative logits (inhibitory scores) or
positive logits (excitatory scores). In the end, the scores are added together to produce a final bag score that is passed through a softmax, returning the final probability for each class.
By comparing the attention scores produced by the two models, we noticed that the use of the LeakyReLU activation function on the attention layer of the AdMIL architecture produced narrower attention scores, which might not be ideal, as it makes the model focus on a limited part of the input. We decided to experiment on an additional model that uses the architecture of AdMIL, but with the attention layer of AMIL.
Authors:
(1) Martim Afonso, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, Lisbon, 1049-001, Portugal;
(2) Praphulla M.S. Bhawsar, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(3) Monjoy Saha, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(4) Jonas S. Almeida, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, 20850, Maryland, USA;
(5) Arlindo L. Oliveira, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, Lisbon, 1049-001, Portugal and INESC-ID, R. Alves Redol 9, Lisbon, 1000-029, Portugal.
This paper is