Table of Links
- Abstract and Introduction
- SylloBio-NLI
- Empirical Evaluation
- Related Work
- Conclusions
- Limitations and References
A. Formalization of the SylloBio-NLI Resource Generation Process
B. Formalization of Tasks 1 and 2
C. Dictionary of gene and pathway membership
D. Domain-specific pipeline for creating NL instances and E Accessing LLMs
F. Experimental Details
G. Evaluation Metrics
H. Prompting LLMs – Zero-shot prompts
I. Prompting LLMs – Few-shot prompts
J. Results: Misaligned Instruction-Response
K. Results: Ambiguous Impact of Distractors on Reasoning
L. Results: Models Prioritize Contextual Knowledge Over Background Knowledge
M Supplementary Figures and N Supplementary Tables
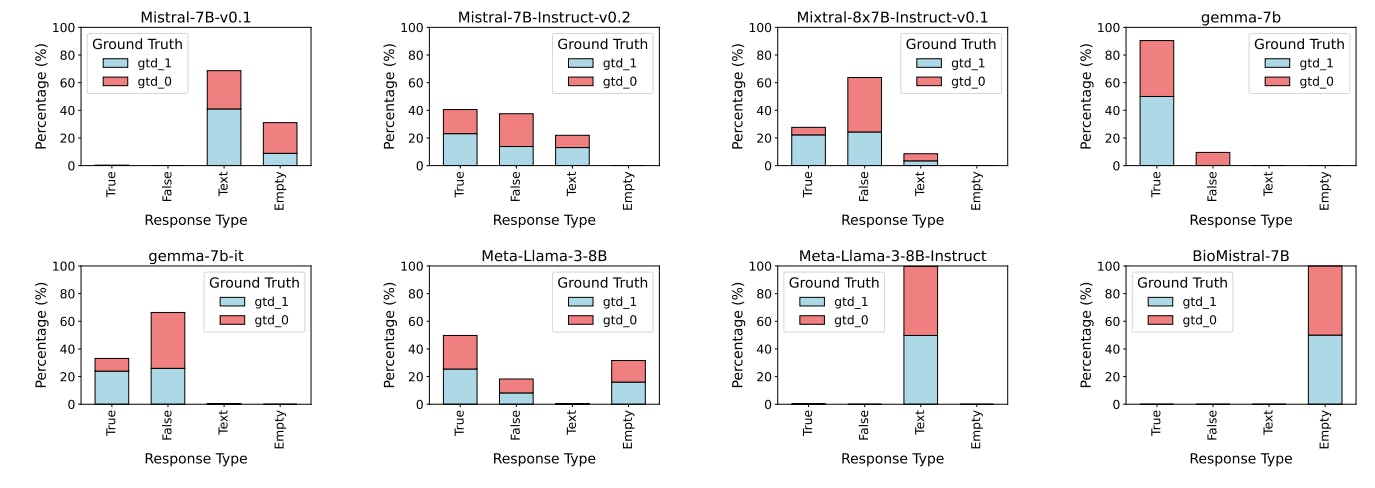
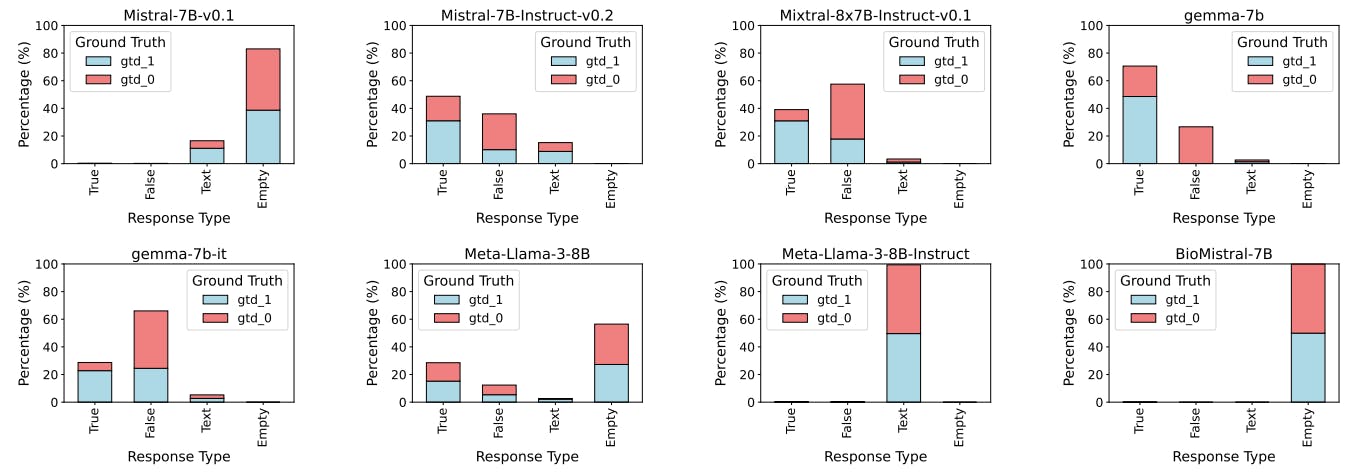
J Results: Misaligned Instruction-Response
We observed four types of text outputs: those aligned with the instruction (regardless of correctness), empty outputs where no text was generated, incorrect text outputs such as repeated prompts or random content, and outputs resembling Chain-of-Thought (CoT) reasoning that, while potentially containing correct reasoning, did not align with the given instructions (Figs. 8-11). We noticed that BioMistral-7B generated empty outputs in 100% of the cases regardless of the specific settings, while Meta-Llama-3-8B exhibits this behaviour for ZS settings in both tasks. We attribute this observation to safety mechanisms applied during pre-training Labrak et al. [2024], suggesting that domain-specific knowledge about human genome pathways is absent in both models. Similarly, Mistral-7B-v0.1 responses simply repeat the prompt text in 88% of the cases in the ZS settings, and 69% of the cases in FS (Table 1). Moreover, CoT outputs including phrases like e.g. “A nice logical puzzle! Let’s break it down step by step…” were particularly common for Meta-Llama-3-8B Instruct, which often ignored the specific instructions to address the task. This behaviour highlights potential biases introduced during instruction-tuning which make the models unable to generalise to domains that are out-of-distribution of the training set.
Authors:
(1) Magdalena Wysocka, National Biomarker Centre, CRUK-MI, Univ. of Manchester, United Kingdom;
(2) Danilo S. Carvalho, National Biomarker Centre, CRUK-MI, Univ. of Manchester, United Kingdom and Department of Computer Science, Univ. of Manchester, United Kingdom;
(3) Oskar Wysocki, National Biomarker Centre, CRUK-MI, Univ. of Manchester, United Kingdom and ited Kingdom 3 I;
(4) Marco Valentino, Idiap Research Institute, Switzerland;
(5) André Freitas, National Biomarker Centre, CRUK-MI, Univ. of Manchester, United Kingdom, Department of Computer Science, Univ. of Manchester, United Kingdom and Idiap Research Institute, Switzerland.