Table of Links
-
Abstract and Introduction
-
System Architecture
2.1 Access via UI or HTTP
2.1.1 GUI
2.2 Input
2.3 Natural Language Processing Pipeline — The Llettuce API
2.3.1 Vector search
2.3.2 LLM
2.3.3 Concept Matches
2.4 Output
-
Case Study: Medication Dataset
3.1 Data Description
3.2 Experimental Design
3.3 Results
3.3.1 Comparison between vector search and Usagi
3.3.2 Comparison with GPT-3
3.4 Conclusions & Acknowledgement
3.5 References
3. Case Study: Medication Dataset
Medication data were obtained from the Health for Life in Singapore (HELIOS) study (IRB approval by Nanyang Technological University: IRB-2016-11-030), a phenotyped longitudinal population cohort study comprising 10,004 multi-ethnic Asian population of Singapore aged 30-85 years (Wang et al., 2024). Participants in the HELIOS study were recruited from the Singapore general population between 2018 and 2022 and underwent extensive clinical, behavioural, molecular and genetic characterisation. With rich baseline data and long-term follow-up through linkage to national health data, the HELIOS study provides a unique and world class resource for biomedical researchers across a wide range of disciplines to understand the aetiology and pathogenesis of diverse disease outcomes in Asia, with potential to improve health and advance healthcare for Asian populations.
To facilitate scalable and collaborative research, the HELIOS study implements the OMOP-CDM. However, mapping medication data to OMOP concepts poses significant challenges, primarily due to the complexities involved in standardising medication names. In the HELIOS study, medication data were self-reported and manually entered via nurse-administered questionnaires, therefore, medications with brand name, abbreviations, typographic misspellings or phonetic errors, or combined medications could be recorded. All of these sources of imprecision make mapping to a controlled medical vocabulary more difficult and require significant manual data cleaning.
3.1 Data Description
The first 400 examples from the medication dataset were selected for our experiments and comparison. For each instance, the best OMOP concept, as well as a broader set of concepts which could match the informal name were compiled by human annotation.
For example, for “Memantine HCl”, the best OMOP concept is “memantine hydrochloride”, although “memantine” is another acceptable answer. For a branded medication, the concept representing the branded product is the most appropriate OMOP concept. The generic ingredient names can be included in a broader set of acceptable concepts, provided all the ingredients are listed within the concept. For example, for “cocodamol capsule”, “Acetaminophen / Codeine Oral Capsule [Co-codamol]” would be the best match, but “acetaminophen/codeine” would be accepted as a broader definition. This also further 297 illustrates the challenges with mapping and the potential uncertainties that the problem presents.
Of the 400 examples, 25 were graded as “Not Parsable”. These were either formulations containing several ingredients where the formulation has no concept in the OMOP CDM, e.g. “lipesco”, which contains lipoic acid and four vitamins and is not in the OMOP CDM; or where the name could not be resolved, e.g. “Hollister (gout)”.
3.2 Experimental Design
The data instances were run through the vector search and LLM portions of the pipeline and compared with the human annotations. The top 5 results from the vector search were used. Responses were assessed by:
-
Whether the input is an exact match to an OMOP concept
-
Whether the correct OMOP concept is in the result of the vector search
-
Whether the LLM provides the correct answer
-
If the answer was incorrect, whether it is a relevant OMOP concept
The same examples were used as input for Usagi and vector search. For each example and both methods, the top 5 results were taken and each response was classified by whether the correct mapping or a relevant mapping was found.
3.3 Results
Table 1 describes the results of comparing Usagi with Llettuce’s vector search. The number of results with at least one relevant concept in the top 5 was very similar between the methods (68% for both). However, Llettuce outperformed Usagi in returning the correct concept in the top 5 (44% for Usagi, 54% for Llettuce).
3.3.1 Comparison between vector search and Usagi
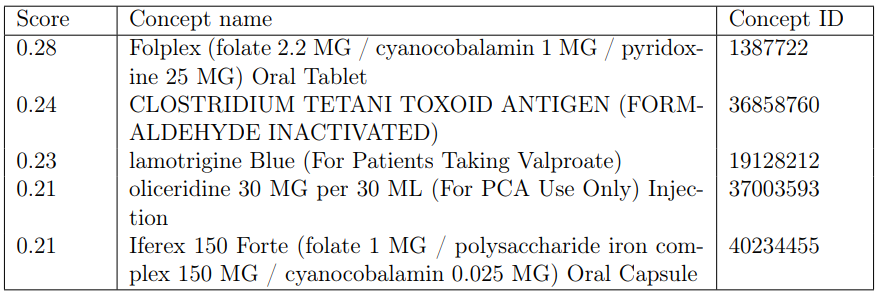
Usagi performs well when used to find concepts where the input has a typographical error. Its shortcomings can be illustrated by how it responds to various descriptions of the mometasone furoate nasal spray, “nasonex”. In the examples, dosage information, such as “Nasonex (for each nostril)” produces the output shown in Table 2 for the top five results.
3.3.2 Comparison with GPT-3
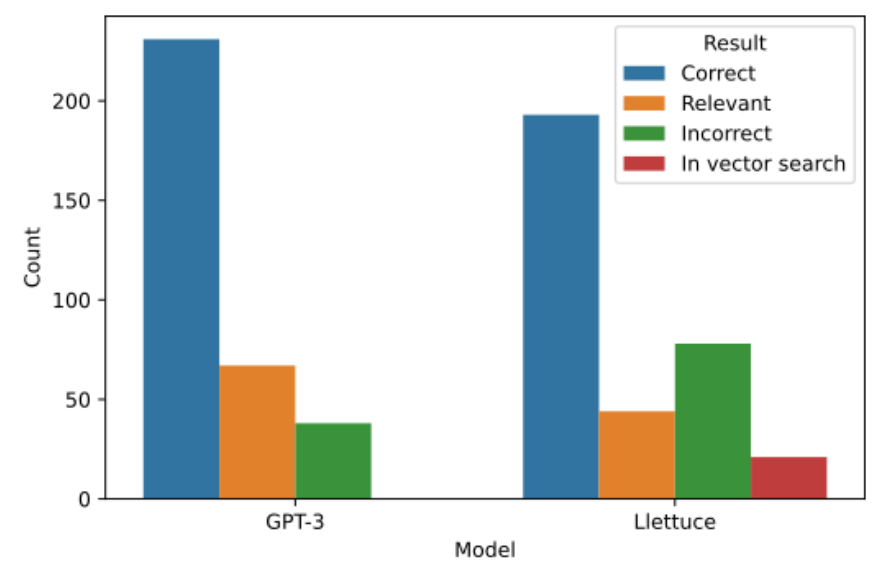
Of the 336 examples where the input was parsable into an OMOP concept, and the input was not an exact match to an OMOP concept, Llettuce could correctly identify 193, or 48.25%. GPT-3 could correctly identify 57.75%. Both provided inexact but matching concepts, 44 (11%) for Llettuce and 67 (16.75%) for GPT-3. The top 5 vector matches 329 retrieved the correct concept for 21 of the 99 inputs incorrectly answered by Llettuce. 232 informal names could be directly mapped onto the best available OMOP concept (if
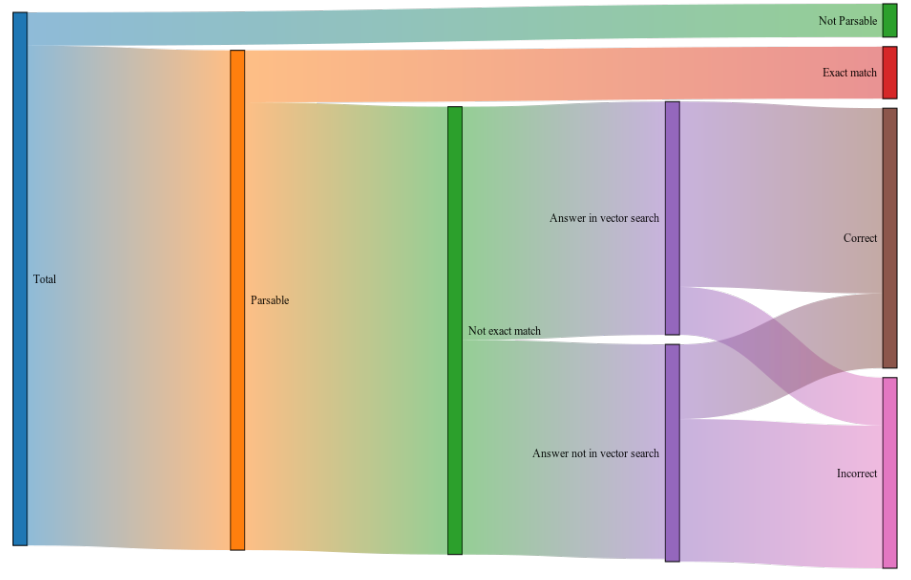

exact matches are included). Of the remaining concepts, 78 had no output that neither included the correct concept nor produced a relevant OMOP concept. Llettuce’s pipeline does not perform as well as GPT-3, which is only absolutely incorrect on 38 names. However, it achieves this run locally on consumer hardware, using a much smaller model and preserving confidentiality.
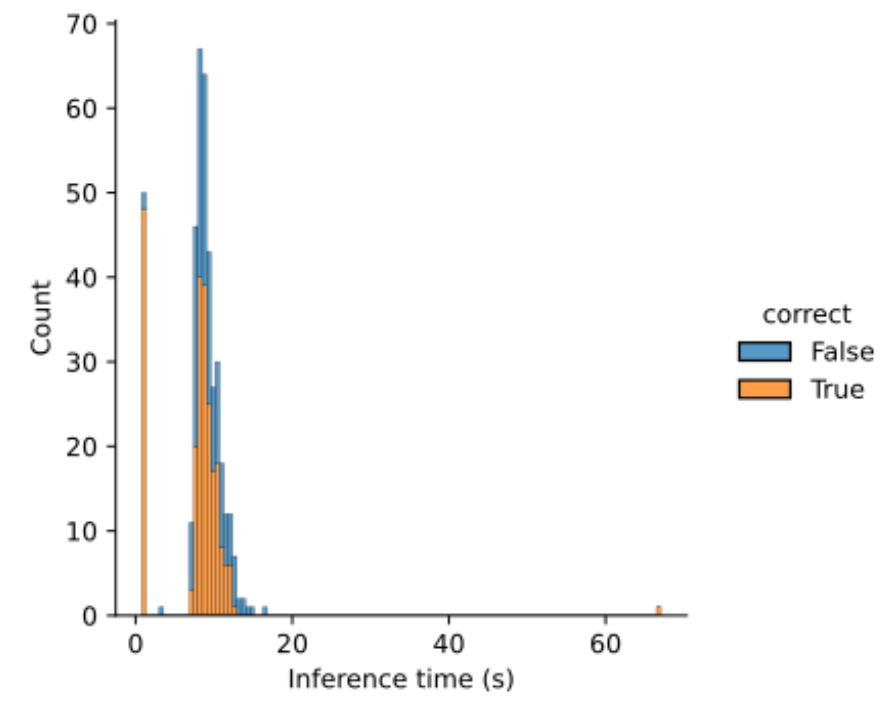
The time taken to run the Llettuce pipeline on 400 concepts was 55 minutes, 15 seconds, using a 2.8GHz quad-core Intel i7 CPU, 16 Gb RAM. The median time to run inference was 8.7 seconds.
3.4. Conclusions
Llettuce demonstrates the possibilities of using deep-learning approaches to map data to OMOP concepts. Combining vector search with a large language model results in comparable performance with the larger GPT-3 model. This shows that the advantages of neural-network based natural language processing can be leveraged to produce medical 344 encodings, even in a setting where confidentiality is essential.
The comparison with string matching methods is also informative. String matching cannot learn the salience of different parts of the string. In the example above, the part of the string “(for each nostril)”, as it is longer, is treated as more important; the algorithm doesn’t know to ignore that part. By contrast, Llettuce’s vector search correctly includes Nasonex in almost all of its inputs, and correctly identifies the active ingredient. It should be noted that in this version of Llettuce only the RxNorm vocabulary was vectorised, where Usagi also used the RxNorm extension. This dataset is also one at which Usagi is relatively good, as it mostly involves extracting a single word, or correcting typographical errors. Anecdotally, Usagi performs worse on other tasks, where the input is longer and semantics are more important. This is where vector search is likely to perform far better. Crucially, an embedding model is trainable, where string comparison is not.
Optimisations will be possible in later versions. The models used for both embeddings and text generation are general purpose models (bge-small-en-v1.5 and Llama-3.1-8B respectively). Existing specialist models either fine-tuned or trained ab initio (Gu et al., 2020) on biomedical literature will be tested for performance on Llettuce tasks. Further development will come from fine-tuned models developed in-house. Our local deployment of Llettuce will implement data collection and record prompts and responses, alongside the final mapping made. This data will be used to fine-tune the models used. It’s important to363 emphasise that this data collection will be strictly limited to our specific local deployment of the tool. The publicly available version will not collect any user data or interactions, 365 maintaining the confidentiality and privacy of health information processed by other users.
Funding
This research was funded by the NIHR Nottingham Biomedical Research Centre.
Data Availability
Data access requests can be submitted to the HELIOS Data Access Committee by emailing [email protected] for details.
Acknowledgments
The authors thank those people or institutions that have helped you in the preparation of the manuscript.
3.5 References
Appleby, P., Masood, E., Milligan, G., Macdonald, C., Quinlan, P., & Cole, C. Carrot-cdm: An open-source tool for transforming data for federated discovery in health research [Research Software Engineering Conference 2023, RSECON23 ; Conference date: 04-09-2023 Through 07-09-2023]. English. In: In Carrot-cdm: An open-source tool for transforming data for federated discovery in health research. Research Software Engineering Conference 2023, RSECON23 ; Conference date: 04-09-2023 Through 07-09-2023. 2023, September. https://doi.org/10.5281/zenodo.10707025
Bayer, M. (2012). Sqlalchemy (A. Brown & G. Wilson, Eds.). http://aosabook.org/en/sqlalchemy.html
Bengio, Y., Ducharme, R., Vincent, P., & Jauvin, C. (2003). A Neural Probabilistic Language Model. Journal of Machine Learning Research, 3.
Brown, T. B., Mann, B., Ryder, N., Subbiah, M., Kaplan, J., Dhariwal, P., Neelakantan, A., Shyam, P., Sastry, G., Askell, A., Agarwal, S., Herbert-Voss, A., Krueger, G., Henighan, T., Child, R., Ramesh, A., Ziegler, D. M., Wu, J., Winter, C., . . . Amodei, D. (2020, July 22). Language Models are Few-Shot Learners. arXiv: 2005.14165 [cs]. https://doi.org/10.48550/arXiv.2005.14165
Cholan, R. A., Pappas, G., Rehwoldt, G., Sills, A. K., Korte, E. D., Appleton, I. K., Scott, N. M., Rubinstein, W. S., Brenner, S. A., Merrick, R., Hadden, W. C., Campbell, K. E., & Waters, M. S. (2022). Encoding laboratory testing data: Case studies of the national implementation of hhs requirements and related standards in five laboratories. Journal of the American Medical Informatics Association, 29(8), 1372–1380. https://doi.org/10.1093/jamia/ocac072
Cox, S., Masood, E., Panagi, V., Macdonald, C., Milligan, G., Horban, S., Santos, R., Hall, C., Lea, D., Tarr, S., Mumtaz, S., Akashili, E., Rae, A., Cole, C., Sheikh, A., Jefferson, E., & Quinlan, P. R. (2024). Improving the quality, speed and transparency of curating data to the observational medical outcomes partnership (OMOP) common data model using the carrot tool. JMIR Preprints. https://doi.org/10.2196/preprints.60917
deepset GmbH. (2024). Haystack: Neural question answering at scale [Accessed: 16-08-2024].
Deng, H., Zhou, Q., Zhang, Z., Zhou, T., Lin, X., Xia, Y., Fan, L., & Liu, S. (2024). The current status and prospects of large language models in medical application and research. Chinese Journal of Academic Radiology. https://doi.org/10.1007/s42058- 024-00164-x
Dettmers, T., & Zettlemoyer, L. (2023, February 27). The case for 4-bit precision: K-bit Inference Scaling Laws. arXiv: 2212.09720 [cs]. https://doi.org/10.48550/arXiv.2212.09720
Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2019, May 24). BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv: 1810.04805 [cs]. https://doi.org/10.48550/arXiv.1810.04805
Dubey, A., Jauhri, A., Pandey, A., Kadian, A., Al-Dahle, A., Letman, A., Mathur, A., Schelten, A., Yang, A., Fan, A., Goyal, A., Hartshorn, A., Yang, A., Mitra, A., Sravankumar, A., Korenev, A., Hinsvark, A., Rao, A., Zhang, A., . . . Zhao, Z. (2024, August 15). The Llama 3 Herd of Models. arXiv: 2407.21783 [cs]. https://doi.org/10.48550/arXiv.2407.21783
F., H., J., H., K., T., A., H., M.J., M., T.W.R., B., J., Y., J., D., A., W., S., E.-J., & W.K., G. (2022). Data consistency in the english hospital episodes statistics database. BMJ Health Care Inform, 29(1), e100633. https://doi.org/10.1136/bmjhci-2022-100633
Gerganov, G., et al. (2024). Llama.cpp [Accessed: 19-08-2024]. https://github.com/ggerganov/llama.cpp
Gu, Y., Tinn, R., Cheng, H., Lucas, M., Usuyama, N., Liu, X., Naumann, T., Gao, J., & Poon, H. (2020). Domain-specific language model pretraining for biomedical natural language processing.
Jacob, B., Kligys, S., Chen, B., Zhu, M., Tang, M., Howard, A., Adam, H., & Kalenichenko, D. (2017, December 15). Quantization and Training of Neural Networks for Efficient Integer-Arithmetic-Only Inference. arXiv: 1712 . 05877 [cs, stat]. https://doi.org/10.48550/arXiv.1712.05877
Kong, A., Zhao, S., Chen, H., Li, Q., Qin, Y., Sun, R., Zhou, X., Wang, E., & Dong, X. (2024, March). Better Zero-Shot Reasoning with Role-Play Prompting.
Lewis, P., Perez, E., Piktus, A., Petroni, F., Karpukhin, V., Goyal, N., Küttler, H., Lewis, M., Yih, W.-t., Rocktäschel, T., Riedel, S., & Kiela, D. (2021, April 12). Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks. arXiv: 2005. 11401 [cs]. https://doi.org/10.48550/arXiv.2005.11401
Meta-llama/llama-recipes. (2024, August 19). Retrieved August 19, 2024, from https://github.com/meta-llama/llama-recipes
Nazi, Z. A., & Peng, W. (2024). Large language models in healthcare and medical domain: A review. Informatics, 11(3), 57. https://doi.org/10.3390/informatics11030057
OHDSI. (2021). (observational health data sciences and informatics), Usagi documentation [Accessed: 13-08-2024]. https://ohdsi.github.io/Usagi/
OHDSI. (2024a). Athena: Standardized vocabularies [Accessed: 16-08-2024]. https://athena.ohdsi.org/search-terms/start
OHDSI. (2024b). Data standardization [Accessed: September 2024]. https://www.ohdsi.org/data-standardization/
OpenAI. (2024). Chatgpt: Language model [Accessed: 2024-08-16]. https://chat.openai.com/
Qdrant/fastembed. (2024, August 19). Retrieved August 19, 2024, from https://github.com/qdrant/fastembed
Ramírez, S. (2024). Fastapi [Accessed: 19-08-2024]. https://fastapi.tiangolo.com
Reimers, N., & Gurevych, I. (2019, August 27). Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks. arXiv: 1908. 10084 [cs]. https://doi.org/10.48550/arXiv.1908.10084
Streamlit. (2024). Streamlit: The fastest way to build and share data apps [Accessed: 16-08-2024]. https://streamlit.io
Wang, X., Mina, T., Sadhu, N., Jain, P. R., Ng, H. K., Low, D. Y., Tay, D., Tong, T. Y. Y., Choo, W.-L., Kerk, S. K., Low, G. L., Team, T. H. S., Lam, B. C. C., Dalan, R., Wanseicheong, G., Yew, Y. W., Leow, E.-J., Brage, S., Michelotti, G. A., . . . Chambers, J. C. (2024, May 24). The Health for Life in Singapore (HELIOS) Study: Delivering Precision Medicine research for Asian populations. https://doi.org/10.1101/2024.05.14.24307259
Wilkinson, M., Dumontier, M., Aalbersberg, I., et al. (2016). The fair guiding principles for scientific data management and stewardship. Scientific Data, 3, 160018. https://doi.org/10.1038/sdata.2016.18
:::info
Authors:
(1) James Mitchell-White, Centre for Health Informatics, School of Medicine, The University of Nottingham, Digital Research Service, The University of Nottingham, and NIHR Nottingham Biomedical Research Centre;
(2) Reza Omdivar, Digital Research Service, The University of Nottingham, and NIHR Nottingham Biomedical Research Centre;
(3) Esmond Urwin, Centre for Health Informatics, School of Medicine, The University of Nottingham and NIHR Nottingham Biomedical Research Centre;
(4) Karthikeyan Sivakumar, Digital Research Service, The University of Nottingham;
(5) Ruizhe Li, NIHR Nottingham Biomedical Research Centre and School of Computer Science, The University of Nottingham;
(6) Andy Rae, Centre for Health Informatics, School of Medicine, The University of Nottingham;
(7) Xiaoyan Wang, Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore;
(8) Theresia Mina, Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore;
(9) John Chambers, Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore and Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, United Kingdom;
(10) Grazziela Figueredo, Centre for Health Informatics, School of Medicine, The University of Nottingham and NIHR Nottingham Biomedical Research Centre;
(11) Philip R Quinlan, Centre for Health Informatics, School of Medicine, The University of Nottingham.
:::
:::info
This paper is available on arxiv under CC BY 4.0 license.
:::